


ANOVA OV result package
Francesco Iorio, EMBL - European Bioinformatics Institute
result parsing started on 2016-04-08 12:06:52 iorio@ebi.ac.uk
Drug Domain: GDSC1000_paper_set, Analysis Domain: OV specific
Super Analysis Details
Screening Version: v17a - Mar2016 2016-03-31
Drug Domain: GDSC1000_paper_set
Time: 2016-03-31_12-36-37
Machine: MacBook-Pro-6
User: iorio
Total Number of ANOVA tests performed: 10911 (72% of total number of drug/feature combos)
Total number of tested drugs: 224
Total number of tested genomic features (mutated driver genes and copy number altered genomic regions): 57
Total number of screened cell lines: 34
MicroSatellite instability included as factor = TRUE
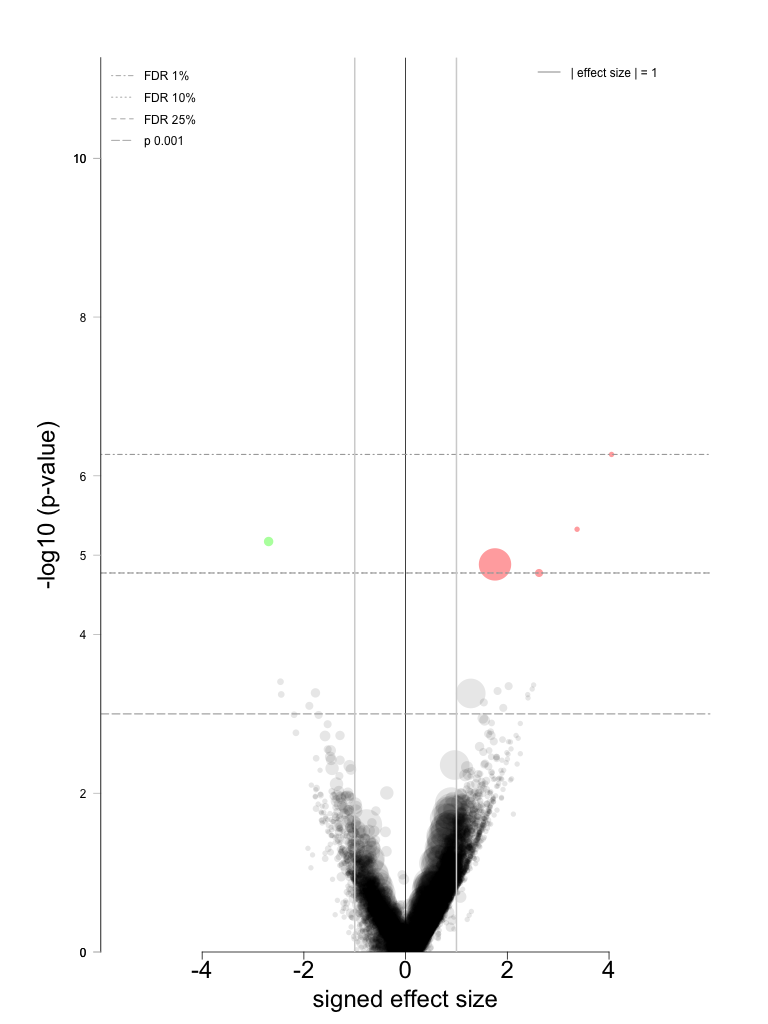
Total number of significant associations: 5 (1 for sensitivity and 4 for resistance)
p-value significance threshold: 0.001, % FDR significance threshold: 25
range of significant p-values: [5.38e-07, 1.68e-05]
range of significant % FDRs: [0.511, 3.18]
Explore all the significant associations
All tested associations with statistical outcomes in an individual tab delimited txt file
ANOVA input data as tab delimited txt file (genomic features and drug response)
CNA segment annotations as tab delimited txt file
Drug annotations as tab delimited txt file
Results summaries
- Comprehensive volcano plots
- Features most frequently associated with drug response:
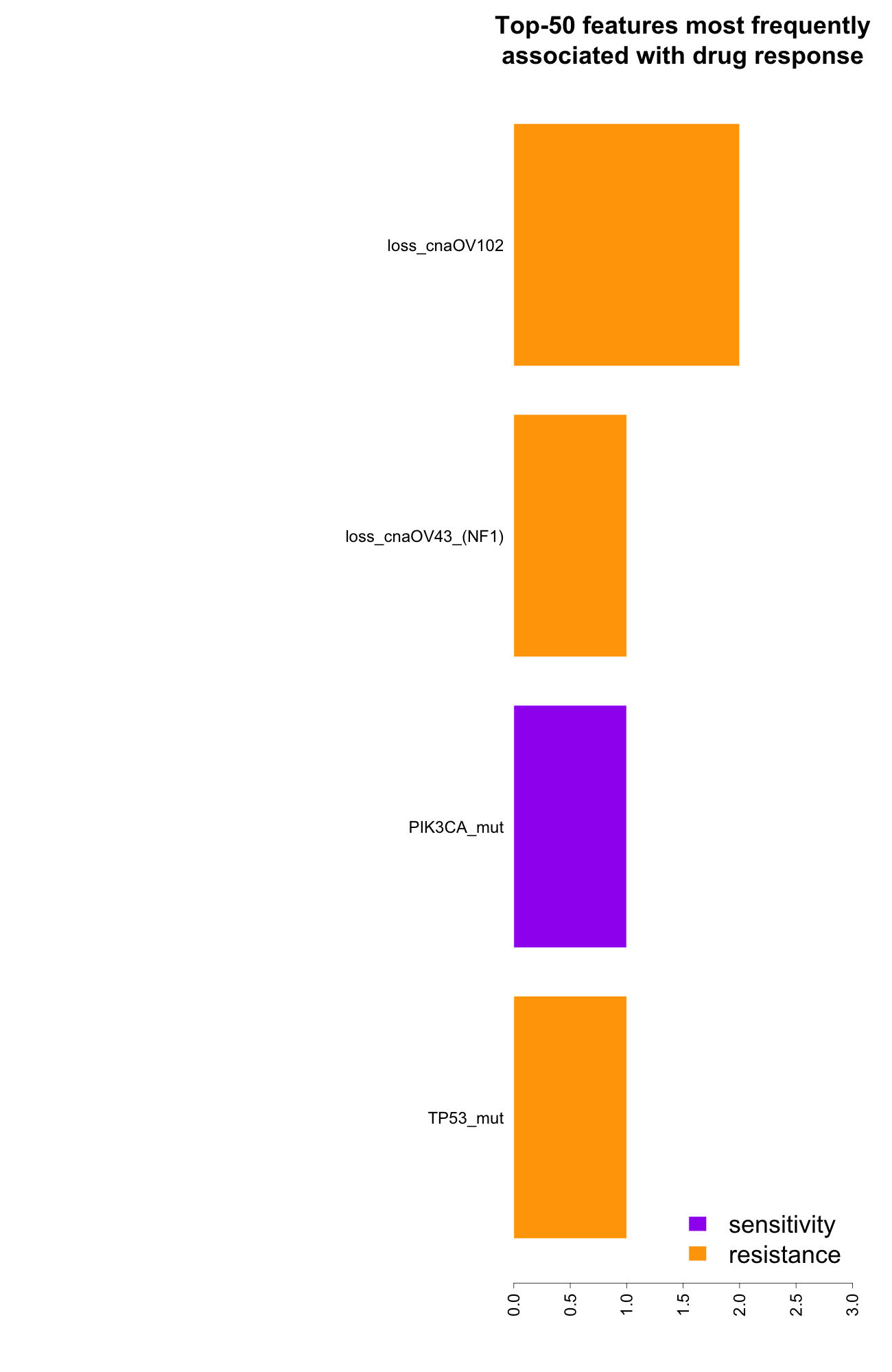
right click on this link to save summary infos for all the features as tab delimited txt file
- Drugs whose response is frequently associated with a feature:
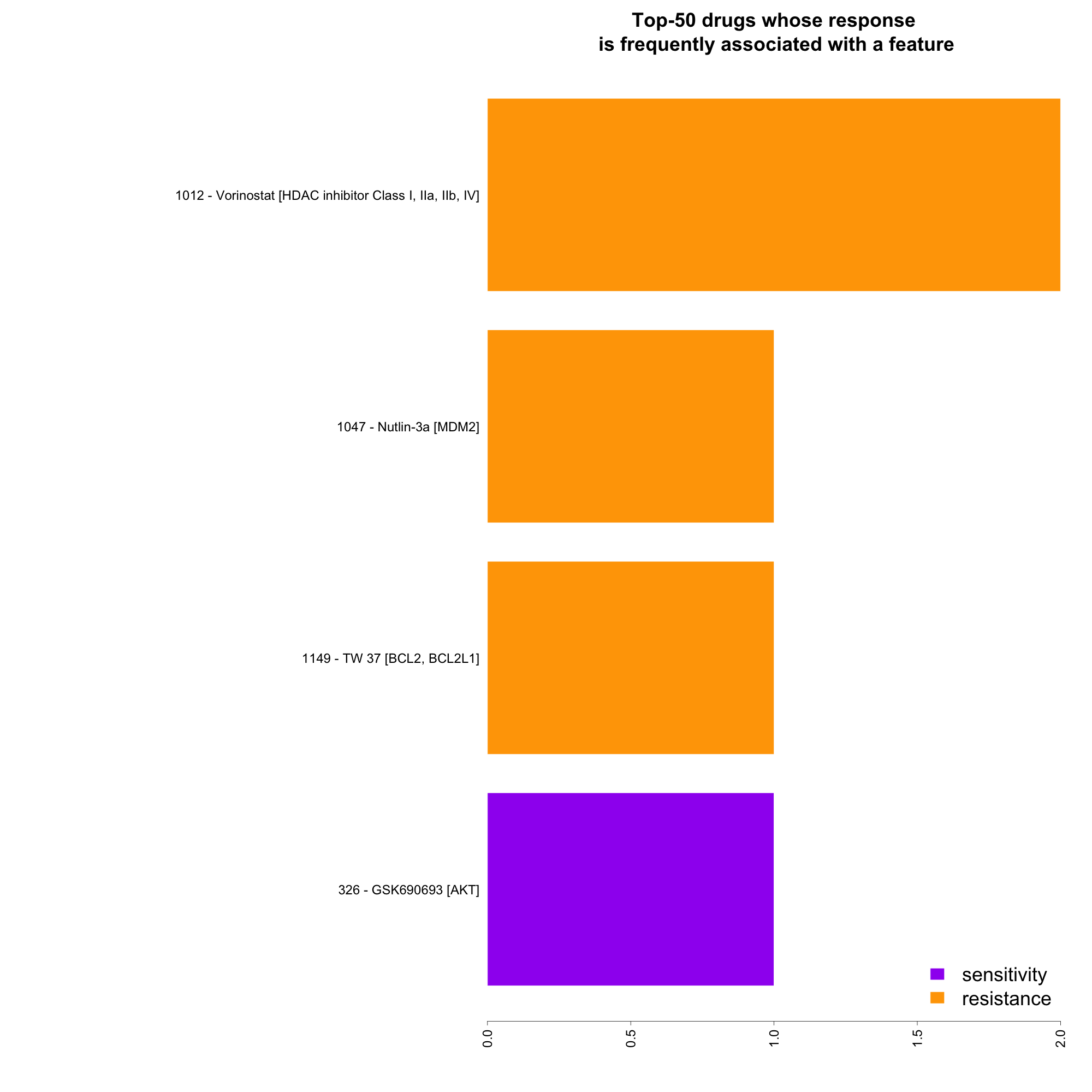
right click on this link to save summary infos for all the features as tab delimited txt file
Drug-wise associations browser
1001 - AICAR
1003 - Camptothecin
1004 - Vinblastine
1005 - Cisplatin
1006 - Cytarabine
1007 - Docetaxel
1008 - Methotrexate
1009 - ATRA
1010 - Gefitinib
1011 - ABT-263
1012 - Vorinostat
1013 - Nilotinib
1014 - RDEA119
1015 - CI-1040
1016 - Temsirolimus
1017 - Olaparib
1018 - ABT-888
1019 - Bosutinib
1020 - Lenalidomide
1021 - Axitinib
1022 - AZD7762
1023 - GW 441756
1024 - CEP-701
1025 - SB 216763
1026 - 17-AAG
1028 - VX-702
1029 - AMG-706
1030 - KU-55933
1031 - Elesclomol
1032 - Afatinib
1033 - Vismodegib
1036 - PLX4720
1037 - BX-795
1038 - NU-7441
1039 - SL 0101-1
1042 - BIRB 0796
1043 - JNK Inhibitor VIII
1046 - 681640
1047 - Nutlin-3a
1049 - PD-173074
1050 - ZM-447439
1052 - RO-3306
1053 - MK-2206
1054 - PD-0332991
1057 - NVP-BEZ235
1058 - GDC0941
1059 - AZD8055
1060 - PD-0325901
1061 - SB590885
1062 - AZD6244
1066 - AZD6482
1067 - CCT007093
1069 - EHT 1864
1072 - BMS-708163
1091 - BMS-536924
1114 - Cetuximab
1129 - PF-4708671
1133 - JNJ-26854165
1142 - HG-5-113-01
1143 - HG-5-88-01
1149 - TW 37
1158 - XMD11-85h
1161 - ZG-10
1164 - XMD8-92
1166 - QL-VIII-58
1170 - CCT018159
1175 - AG-014699
1192 - GSK269962A
1194 - SB-505124
1199 - Tamoxifen
1203 - QL-XII-61
1218 - JQ1
1219 - PFI-1
1230 - IOX2
1236 - UNC0638
1239 - YK 4-279
1241 - CHIR-99021
1242 - (5Z)-7-Oxozeaenol
1243 - piperlongumine
1248 - FK866
1259 - BMN-673
1261 - rTRAIL
1262 - UNC1215
1264 - SGC0946
1268 - XAV 939
133 - Doxorubicin
134 - Etoposide
135 - Gemcitabine
136 - Mitomycin C
1371 - PLX4720 (rescreen)
1372 - Trametinib
1373 - Dabrafenib
1375 - Temozolomide
1377 - Afatinib (rescreen)
1378 - Bleomycin (50 uM)
140 - Vinorelbine
147 - NSC-87877
1494 - SN-38
1495 - AZD-2281
1498 - AZD6244
150 - Bicalutamide
1502 - Bicalutamide
151 - QS11
152 - CP466722
1526 - RDEA119 (rescreen)
1527 - GDC0941 (rescreen)
1529 - MLN4924
153 - Midostaurin
154 - CHIR-99021
155 - AP-24534
156 - AZD6482
157 - JNK-9L
158 - PF-562271
159 - HG-6-64-1
163 - JQ1
164 - JQ12
165 - DMOG
166 - FTI-277
167 - OSU-03012
170 - Shikonin
171 - AKT inhibitor VIII
172 - Embelin
173 - FH535
175 - PAC-1
176 - IPA-3
177 - GSK-650394
178 - BAY 61-3606
179 - 5-Fluorouracil
180 - Thapsigargin
182 - Obatoclax Mesylate
184 - BMS-754807
185 - OSI-906
186 - Bexarotene
190 - Bleomycin
192 - LFM-A13
193 - GW-2580
194 - AUY922
196 - Phenformin
197 - Bryostatin 1
199 - Pazopanib
200 - LAQ824
201 - Epothilone B
202 - GSK-1904529A
203 - BMS-345541
204 - Tipifarnib
205 - BMS-708163
206 - Ruxolitinib
207 - AS601245
208 - SB-715992
211 - TL-2-105
219 - AT-7519
221 - TAK-715
222 - BX-912
223 - ZSTK474
224 - AS605240
225 - Genentech Cpd 10
226 - GSK1070916
228 - KIN001-102
229 - LY317615
230 - GSK429286A
231 - FMK
235 - QL-XII-47
238 - CAL-101
245 - UNC0638
249 - XL-184
252 - WZ3105
253 - XMD14-99
254 - AC220
255 - CP724714
256 - JW-7-24-1
257 - NPK76-II-72-1
258 - STF-62247
260 - NG-25
261 - TL-1-85
262 - VX-11e
263 - FR-180204
265 - Tubastatin A
266 - Zibotentan, ZD4054
268 - YM155
269 - NSC-207895
271 - VNLG/124
272 - AR-42
273 - CUDC-101
274 - PXD101, Belinostat
275 - I-BET
276 - CAY10603
277 - ABT-869
279 - BIX02189
281 - CH5424802
282 - EKB-569
283 - GSK2126458
286 - KIN001-236
287 - KIN001-244
288 - KIN001-055
290 - KIN001-260
291 - KIN001-266
292 - Masitinib
293 - MP470
294 - MPS-1-IN-1
295 - NVP-BHG712
298 - OSI-930
299 - OSI-027
300 - CX-5461
301 - PHA-793887
302 - PI-103
303 - PIK-93
304 - SB52334
305 - TPCA-1
306 - TG101348
308 - XL-880
309 - Y-39983
310 - YM201636
312 - AV-951
326 - GSK690693
328 - SNX-2112
329 - QL-XI-92
330 - XMD13-2
331 - QL-X-138
332 - XMD15-27
333 - T0901317
341 - EX-527
344 - THZ-2-49
345 - KIN001-270
346 - THZ-2-102-1
Feature-wise associations browser
ARID1A_mut
BRCA1_mut
gain_cnaOV18_(CCNE1)
gain_cnaOV19
gain_cnaOV26
gain_cnaOV4_(KRAS)
gain_cnaOV48_(SOX17)
gain_cnaOV49
gain_cnaOV50
gain_cnaOV51_(MYC)
gain_cnaOV72_(ASXL1)
gain_cnaOV94
gain_cnaOV95_(MECOM)
gain_cnaOV96
KRAS_mut
loss_cnaOV1
loss_cnaOV102
loss_cnaOV16, loss_cnaOV17
loss_cnaOV20
loss_cnaOV21, loss_cnaOV22
loss_cnaOV27, loss_cnaOV28, loss_cnaOV29, loss_cnaOV30
loss_cnaOV33_(ZFHX3)
loss_cnaOV34
loss_cnaOV35
loss_cnaOV36
loss_cnaOV38
loss_cnaOV39
loss_cnaOV40
loss_cnaOV41
loss_cnaOV42
loss_cnaOV43_(NF1)
loss_cnaOV44
loss_cnaOV52, loss_cnaOV53
loss_cnaOV54
loss_cnaOV55
loss_cnaOV56
loss_cnaOV57_(ZNRF3)
loss_cnaOV58
loss_cnaOV59
loss_cnaOV61, loss_cnaOV62, loss_cnaOV63
loss_cnaOV64_(ARID1B,MAP3K4,MLLT4)
loss_cnaOV70
loss_cnaOV81_(RB1)
loss_cnaOV82
loss_cnaOV83
loss_cnaOV84
loss_cnaOV85
loss_cnaOV87, loss_cnaOV88
MLL2_mut
NF1_mut
PIK3CA_mut
PIK3R1_mut
PTEN_mut
RB1_mut
SMAD4_mut
SMARCA4_mut
TP53_mut





