ANOVA PANCAN result package
Francesco Iorio, EMBL - European Bioinformatics Institute
result parsing started on 2016-03-31 19:01:41 iorio@ebi.ac.uk
Drug Domain: GDSC1000_paper_set, Analysis Domain: panCancer
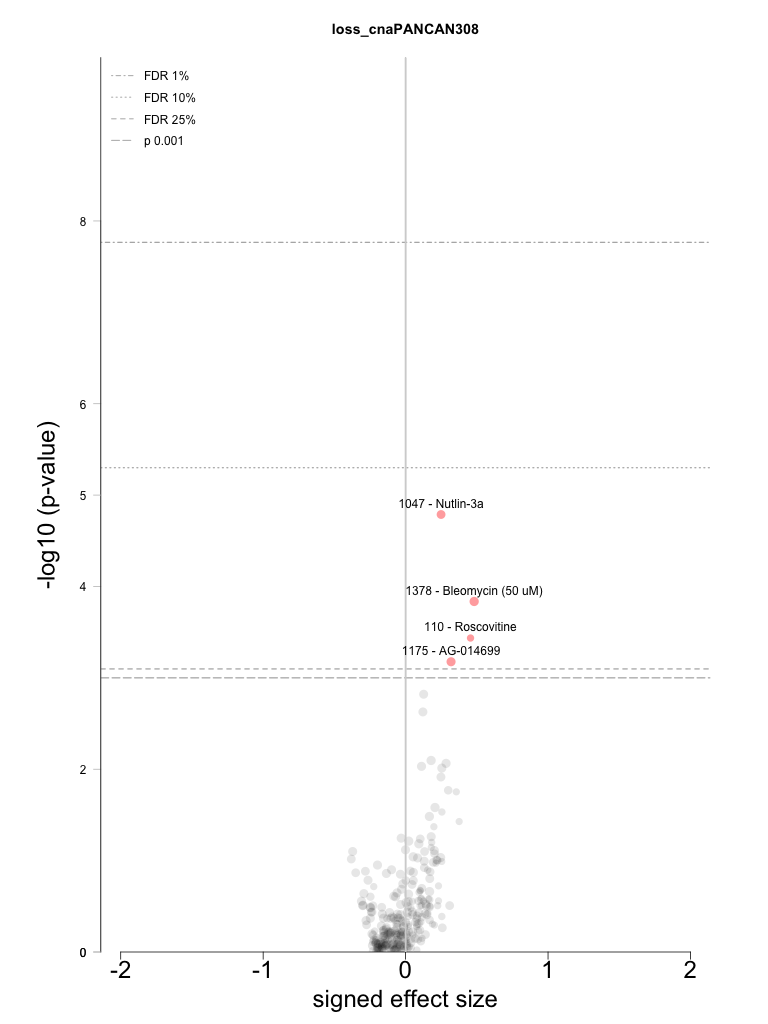
Individual Feature analysis: loss_cnaPANCAN308
Feature details and cell panel numbers
Binary feature equal to 1 for samples having the genomic region(s) cnaPANCAN308 deleted (see genomic region decoding file for further infos on this region)
Number of cell lines positive for this feature: 98
Statistically significant associations identified = 4
| association id |
FEATURE |
Drug id |
Propr |
Public |
Drug Name |
Drug Target |
N FEATURE pos |
N FEATURE neg |
log max Conc tested |
FEATURE neg logIC50 MEAN |
FEATURE pos logIC50 MEAN |
FEATURE deltaMEAN IC50 |
FEATURE IC50 effectSize |
FEATURE neg Glass delta |
FEATURE pos Glass delta |
ANOVA FEATURE pval |
Tissue ANOVA pval |
MSI ANOVA pval |
ANOVA FEATURE FDR % |
a105 |
loss_cnaPANCAN308 |
1047 |
1 |
1 |
Nutlin-3a |
MDM2 |
81 |
765 |
2.08, 1.39 |
3.49 |
3.83 |
0.337 |
0.25 |
0.24 |
0.3 |
1.63e-05 |
5.46e-32 |
6.29e-02 |
2.6 |
a244 |
loss_cnaPANCAN308 |
1378 |
1 |
1 |
Bleomycin (50 uM) |
DNA damage |
91 |
841 |
3.91 |
2.23 |
3.22 |
0.991 |
0.48 |
0.47 |
0.56 . |
1.46e-04 |
5.83e-31 |
3.33e-04 |
9.9 |
a364 |
loss_cnaPANCAN308 |
110 |
1 |
1 |
Roscovitine |
CDKs |
47 |
350 |
2.77 |
3.87 |
4.32 |
0.442 |
0.46 |
0.46 |
0.42 |
3.67e-04 |
2.1e-04 |
8.98e-01 |
17 |
a480 |
loss_cnaPANCAN308 |
1175 |
1 |
1 |
AG-014699 |
PARP1, PARP2 |
87 |
831 |
1.61 |
3.39 |
3.71 |
0.314 |
0.32 |
0.31 |
0.39 |
6.66e-04 |
9.78e-15 |
1.24e-01 |
23 |
All-tests volcano plot