| association id |
FEATURE |
Drug id |
Propr |
Public |
Drug Name |
Drug Target |
N FEATURE pos |
N FEATURE neg |
log max Conc tested |
FEATURE neg logIC50 MEAN |
FEATURE pos logIC50 MEAN |
FEATURE deltaMEAN IC50 |
FEATURE IC50 effectSize |
FEATURE neg Glass delta |
FEATURE pos Glass delta |
ANOVA FEATURE pval |
Tissue ANOVA pval |
MSI ANOVA pval |
ANOVA FEATURE FDR % |
a2 |
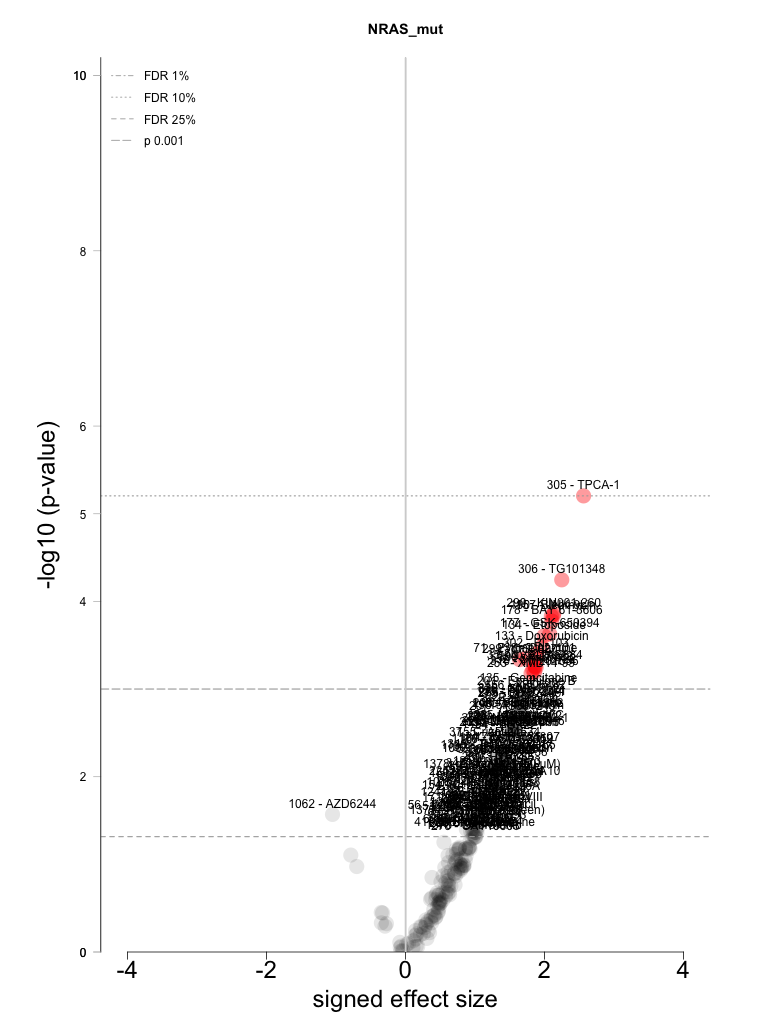
NRAS_mut |
305 |
1 |
1 |
TPCA-1 |
IKK |
7 |
18 |
3 |
-0.0249 |
2.38 |
2.41 |
2.6 ** |
2.4 ** |
3 ** |
6.26e-06 |
NA |
3.58e-03 |
0.38 |
a3 |
NRAS_mut |
306 |
1 |
1 |
TG101348 |
JAK2 |
7 |
19 |
2.33 |
-0.781 |
1.4 |
2.18 |
2.2 ** |
2.1 ** |
2.8 ** |
5.67e-05 |
NA |
3.68e-02 |
2.3 |
a4 |
NRAS_mut |
290 |
1 |
1 |
KIN001-260 |
IKK |
7 |
18 |
3 |
2.33 |
3.96 |
1.64 |
2.1 ** |
2.3 ** |
1.8 * |
1.37e-04 |
NA |
8.03e-02 |
2.9 |
a5 |
NRAS_mut |
190 |
1 |
1 |
Bleomycin |
DNA damage |
7 |
18 |
4.16 |
0.562 |
3.89 |
3.33 |
2.1 ** |
1.9 * |
4.3 ** |
1.46e-04 |
NA |
9.13e-02 |
2.9 |
a6 |
NRAS_mut |
157 |
1 |
1 |
JNK-9L |
JNK |
7 |
19 |
1.63 |
-0.955 |
0.103 |
1.06 |
2.1 ** |
2.2 ** |
1.8 * |
1.47e-04 |
NA |
1.04e-01 |
2.9 |
a7 |
NRAS_mut |
178 |
1 |
1 |
BAY 61-3606 |
SYK |
7 |
18 |
2.08 |
0.491 |
2.86 |
2.36 |
2.1 ** |
2.2 ** |
1.8 * |
1.67e-04 |
NA |
2.85e-01 |
2.9 |
a9 |
NRAS_mut |
177 |
1 |
1 |
GSK-650394 |
SGK3 |
7 |
17 |
2.77 |
1.52 |
4.19 |
2.68 |
2.1 ** |
1.9 * |
3 ** |
2.36e-04 |
NA |
2.39e-01 |
3 |
a10 |
NRAS_mut |
134 |
1 |
1 |
Etoposide |
TOP2 |
7 |
19 |
2.77 |
0.114 |
2.92 |
2.8 |
2 * |
2.1 ** |
1.7 * |
2.47e-04 |
NA |
3.59e-01 |
3 |
a12 |
NRAS_mut |
133 |
1 |
1 |
Doxorubicin |
DNA intercalating |
7 |
18 |
0.0237 |
-2.47 |
-0.24 |
2.23 |
2 * |
2.3 ** |
1.5 * |
3.3e-04 |
NA |
5.1e-01 |
3.2 |
a14 |
NRAS_mut |
302 |
1 |
1 |
PI-103 |
PI3Ka, PRKDC (DNAPK) |
7 |
19 |
2.33 |
-2.24 |
0.595 |
2.84 |
1.9 * |
2.1 ** |
1.5 * |
3.98e-04 |
NA |
1.65e-02 |
3.3 |
a15 |
NRAS_mut |
71 |
1 |
1 |
Pyrimethamine |
Dihydrofolate reductase (DHFR) |
7 |
19 |
3 |
2.78 |
4.84 |
2.06 |
1.7 * |
1.5 * |
4 ** |
4.45e-04 |
NA |
5.23e-01 |
3.3 |
a16 |
NRAS_mut |
238 |
1 |
1 |
CAL-101 |
PI3Kdelta |
7 |
19 |
3.00, 2.33 |
2.09 |
3.99 |
1.9 |
1.9 * |
2 * |
1.7 * |
4.51e-04 |
NA |
5.21e-02 |
3.3 |
a17 |
NRAS_mut |
299 |
1 |
1 |
OSI-027 |
MTORC1/2 |
7 |
19 |
3 |
-1.88 |
0.296 |
2.17 |
1.6 * |
1.9 * |
1.2 * |
4.61e-04 |
NA |
1.06e-04 |
3.3 |
a18 |
NRAS_mut |
35 |
1 |
1 |
NVP-TAE684 |
ALK |
7 |
19 |
0.693 |
-0.537 |
1.46 |
1.99 |
1.9 * |
1.9 * |
1.8 * |
5.48e-04 |
NA |
2.5e-01 |
3.3 |
a19 |
NRAS_mut |
194 |
1 |
1 |
AUY922 |
HSP90 |
7 |
18 |
-1.36 |
-4.14 |
-2.67 |
1.47 |
1.9 * |
2 ** |
1.5 * |
5.57e-04 |
NA |
4.8e-02 |
3.3 |
a21 |
NRAS_mut |
309 |
1 |
1 |
Y-39983 |
ROCK |
7 |
18 |
3 |
2.05 |
4.12 |
2.07 |
1.9 * |
1.8 * |
2.2 ** |
5.71e-04 |
NA |
2.15e-01 |
3.3 |
a22 |
NRAS_mut |
152 |
1 |
1 |
CP466722 |
ATM |
7 |
19 |
2.77 |
0.62 |
2 |
1.38 |
1.8 * |
1.9 * |
1.8 * |
6.04e-04 |
NA |
3.78e-01 |
3.4 |
a23 |
NRAS_mut |
310 |
1 |
1 |
YM201636 |
FYV1 |
7 |
18 |
1.63 |
0.701 |
1.96 |
1.26 |
1.9 * |
2.4 ** |
1.3 * |
6.41e-04 |
NA |
2.02e-01 |
3.4 |
a24 |
NRAS_mut |
253 |
1 |
1 |
XMD14-99 |
EPHB3, CAMK1 |
7 |
19 |
2.33 |
2.65 |
4 |
1.36 |
1.8 * |
2 * |
1.5 * |
6.61e-04 |
NA |
4.73e-01 |
3.4 |