ANOVA PANCAN result package
Francesco Iorio, EMBL - European Bioinformatics Institute
result parsing started on 2016-03-31 19:01:19 iorio@ebi.ac.uk
Drug Domain: GDSC1000_paper_set, Analysis Domain: panCancer
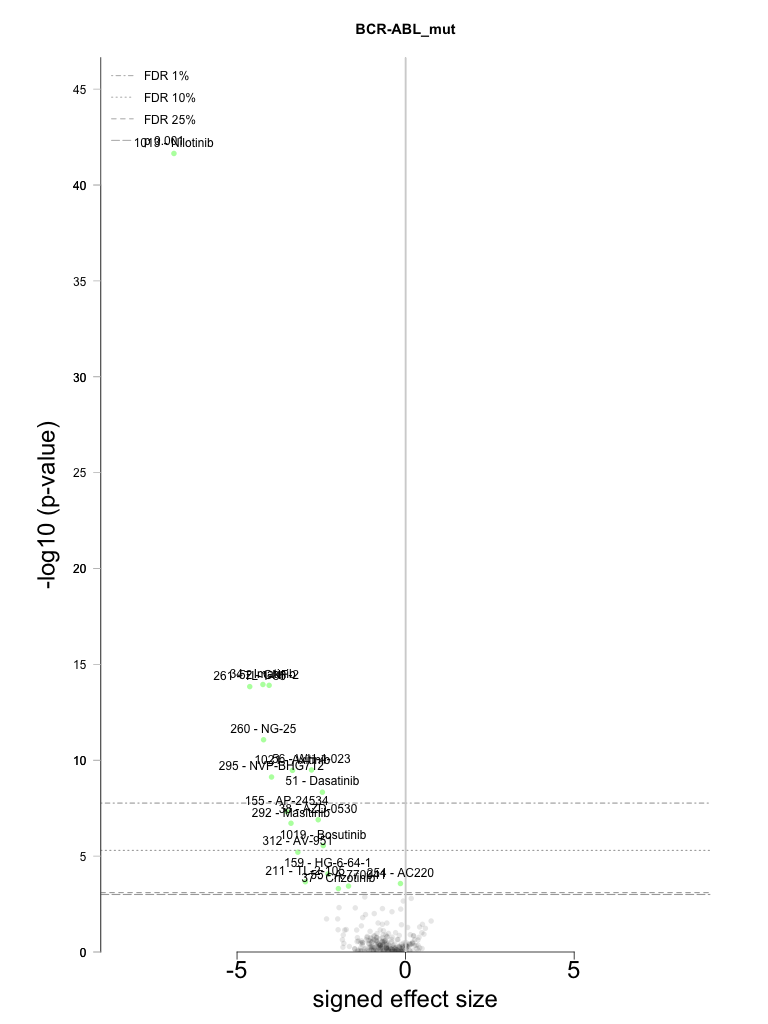
Individual Feature analysis: BCR-ABL_mut
Feature details and cell panel numbers
Binary feature equal to 1 for samples harboring mutations in BCR-ABL
Number of cell lines positive for this feature: 6
Statistically significant associations identified = 19
| association id |
FEATURE |
Drug id |
Propr |
Public |
Drug Name |
Drug Target |
N FEATURE pos |
N FEATURE neg |
log max Conc tested |
FEATURE neg logIC50 MEAN |
FEATURE pos logIC50 MEAN |
FEATURE deltaMEAN IC50 |
FEATURE IC50 effectSize |
FEATURE neg Glass delta |
FEATURE pos Glass delta |
ANOVA FEATURE pval |
Tissue ANOVA pval |
MSI ANOVA pval |
ANOVA FEATURE FDR % |
a3 |
BCR-ABL_mut |
1013 |
1 |
1 |
Nilotinib |
ABL |
6 |
794 |
0.693 |
2.71 |
-5.32 |
-8.03 |
6.9 ** |
6.9 ** |
14 ** |
2.26e-42 |
3.51e-47 |
1.61e-01 |
1.2e-35 |
a7 |
BCR-ABL_mut |
34 |
1 |
1 |
Imatinib |
ABL, KIT, PDGFR |
5 |
403 |
0.693 |
2.79 |
-1.31 |
-4.1 |
4.2 ** |
4.3 ** |
1.9 * |
1.12e-14 |
1.9e-11 |
5.32e-01 |
2.6e-08 |
a8 |
BCR-ABL_mut |
52 |
1 |
1 |
GNF-2 |
ABL [T315I] |
5 |
393 |
0.247 |
2.4 |
-0.761 |
-3.16 |
4 ** |
4.2 ** |
1.7 * |
1.24e-14 |
6.43e-06 |
8.71e-01 |
2.6e-08 |
a9 |
BCR-ABL_mut |
261 |
1 |
1 |
TL-1-85 |
MAP3K7 (TAK1) |
5 |
914 |
2.33 |
3.21 |
-4 |
-7.21 |
4.6 ** |
4.6 ** |
7.2 ** |
1.44e-14 |
1.46e-78 |
5.64e-01 |
2.6e-08 |
a11 |
BCR-ABL_mut |
260 |
1 |
1 |
NG-25 |
MAP3K7 (TAK1) |
5 |
914 |
2.33 |
2.67 |
-4.22 |
-6.89 |
4.2 ** |
4.2 ** |
13 ** |
8.52e-12 |
5.68e-63 |
9.08e-01 |
1.3e-05 |
a18 |
BCR-ABL_mut |
56 |
1 |
1 |
WH-4-023 |
SRC family, ABL |
5 |
393 |
1.63 |
2.17 |
-3.46 |
-5.64 |
2.8 ** |
2.8 ** |
2.1 ** |
3.21e-10 |
6.47e-11 |
5.36e-01 |
0.00029 |
a19 |
BCR-ABL_mut |
1021 |
1 |
1 |
Axitinib |
PDGFR, KIT, VEGFR |
6 |
838 |
0.693 |
1.99 |
-2.17 |
-4.16 |
3.4 ** |
3.3 ** |
11 ** |
3.37e-10 |
4.57e-22 |
9.72e-01 |
0.00029 |
a24 |
BCR-ABL_mut |
295 |
1 |
1 |
NVP-BHG712 |
EPHB4 |
5 |
914 |
2.33 |
3.03 |
-3.32 |
-6.35 |
4 ** |
4 ** |
13 ** |
7.49e-10 |
3.82e-67 |
4.71e-01 |
0.00052 |
a28 |
BCR-ABL_mut |
51 |
1 |
1 |
Dasatinib |
ABL, SRC, KIT, PDGFR |
5 |
392 |
0.247 |
0.331 |
-5.51 |
-5.84 |
2.5 ** |
2.5 ** |
3.1 ** |
4.63e-09 |
6.05e-16 |
4.81e-01 |
0.0027 |
a36 |
BCR-ABL_mut |
155 |
1 |
1 |
AP-24534 |
ABL |
5 |
876 |
-0.669 |
0.437 |
-5.13 |
-5.57 |
3.5 ** |
3.5 ** |
1.9 * |
4.73e-08 |
7.17e-38 |
9.05e-01 |
0.021 |
a44 |
BCR-ABL_mut |
38 |
1 |
1 |
AZD-0530 |
SRC, ABL1 |
5 |
403 |
0.693 |
2.2 |
-0.794 |
-2.99 |
2.6 ** |
2.6 ** |
2.4 ** |
1.27e-07 |
7.54e-06 |
6.34e-01 |
0.048 |
a48 |
BCR-ABL_mut |
292 |
1 |
1 |
Masitinib |
KIT |
5 |
915 |
3 |
3.62 |
-0.696 |
-4.32 |
3.4 ** |
3.4 ** |
2.6 ** |
1.93e-07 |
8.32e-51 |
2.25e-01 |
0.066 |
a69 |
BCR-ABL_mut |
1019 |
1 |
1 |
Bosutinib |
SRC, ABL, TEC |
6 |
842 |
0.693 |
1.77 |
-1.56 |
-3.33 |
2.4 ** |
2.4 ** |
19 ** |
2.82e-06 |
5.4e-28 |
1.1e-01 |
0.67 |
a87 |
BCR-ABL_mut |
312 |
1 |
1 |
AV-951 |
VEGFR |
5 |
914 |
-2.06 |
0.296 |
-2.15 |
-2.45 |
3.2 ** |
3.2 ** |
2.1 ** |
6.23e-06 |
4.03e-52 |
9.64e-01 |
1.2 |
a201 |
BCR-ABL_mut |
159 |
1 |
1 |
HG-6-64-1 |
BRAFV600E, TAK, MAP4K5 |
5 |
872 |
1.63 |
1.02 |
-2.76 |
-3.78 |
2.3 ** |
2.3 ** |
3 ** |
8.53e-05 |
6.53e-26 |
1.34e-01 |
7 |
a290 |
BCR-ABL_mut |
211 |
1 |
1 |
TL-2-105 |
CRAF |
5 |
919 |
2.33, 1.63 |
3.86 |
-0.0316 |
-3.89 |
3 ** |
3 ** |
2.4 ** |
2.17e-04 |
1.08e-68 |
7.35e-01 |
12 |
a308 |
BCR-ABL_mut |
254 |
1 |
1 |
AC220 |
FLT3 |
5 |
915 |
0.0237 |
2.13 |
1.97 |
-0.162 |
0.15 |
0.15 |
0.24 |
2.69e-04 |
2.17e-47 |
9.4e-01 |
14 |
a360 |
BCR-ABL_mut |
55 |
1 |
1 |
A-770041 |
SRC family |
5 |
396 |
1.63 |
2.1 |
-0.873 |
-2.97 |
1.7 * |
1.7 * |
2.7 ** |
3.65e-04 |
2.34e-07 |
5.94e-01 |
17 |
a412 |
BCR-ABL_mut |
37 |
1 |
1 |
Crizotinib |
MET, ALK |
5 |
402 |
0.693 |
2.41 |
0.187 |
-2.22 |
2 * |
2 * |
2.3 ** |
4.96e-04 |
4.97e-11 |
6.04e-01 |
20 |
All-tests volcano plot