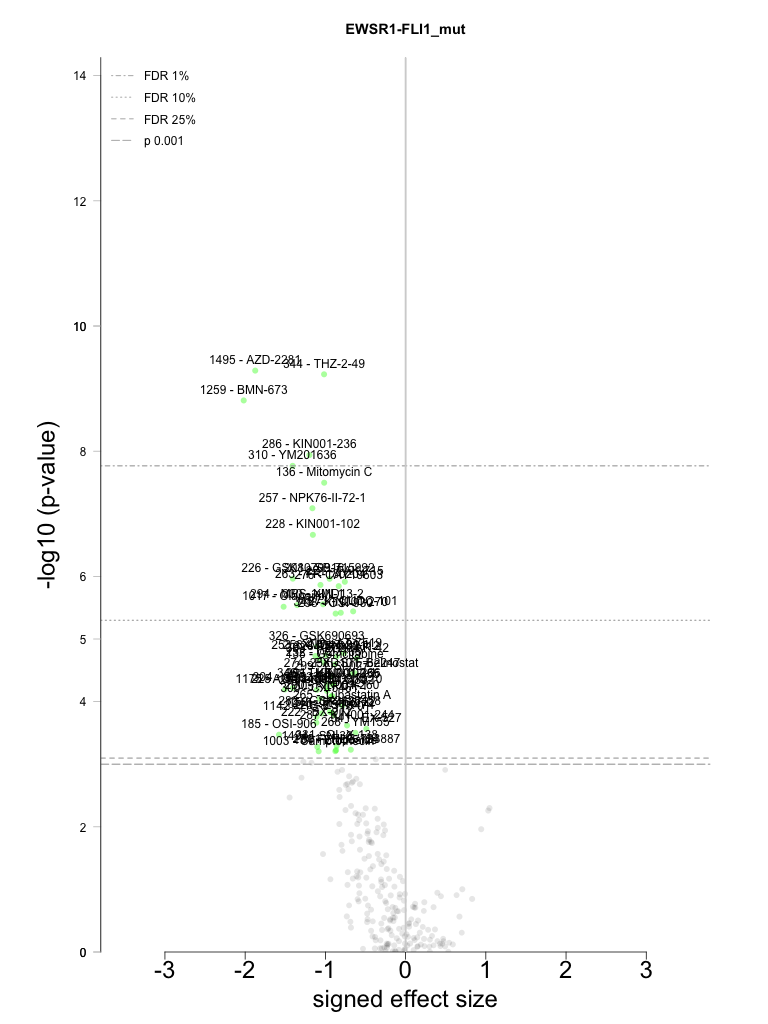
| association id |
FEATURE |
Drug id |
Propr |
Public |
Drug Name |
Drug Target |
N FEATURE pos |
N FEATURE neg |
log max Conc tested |
FEATURE neg logIC50 MEAN |
FEATURE pos logIC50 MEAN |
FEATURE deltaMEAN IC50 |
FEATURE IC50 effectSize |
FEATURE neg Glass delta |
FEATURE pos Glass delta |
ANOVA FEATURE pval |
Tissue ANOVA pval |
MSI ANOVA pval |
ANOVA FEATURE FDR % |
a21 |
EWSR1-FLI1_mut |
1495 |
1 |
1 |
AZD-2281 |
PARP1, PARP2 |
18 |
885 |
2.3 |
3.66 |
1.75 |
-1.9 |
1.9 * |
1.9 * |
1.6 * |
5.15e-10 |
1.8e-17 |
1.18e-03 |
0.00041 |
a23 |
EWSR1-FLI1_mut |
344 |
1 |
1 |
THZ-2-49 |
CDK9 |
17 |
899 |
3 |
2.71 |
0.387 |
-2.33 |
1 * |
1 * |
1.1 * |
5.91e-10 |
2.89e-72 |
2.9e-01 |
0.00042 |
a26 |
EWSR1-FLI1_mut |
1259 |
1 |
1 |
BMN-673 |
PARP1 |
18 |
894 |
2.3 |
2.18 |
-1.48 |
-3.66 |
2 ** |
2 ** |
1.5 * |
1.54e-09 |
4.9e-19 |
1.93e-03 |
0.00098 |
a30 |
EWSR1-FLI1_mut |
286 |
1 |
1 |
KIN001-236 |
TIE2 |
17 |
902 |
2.33 |
4.28 |
3.09 |
-1.18 |
1.2 * |
1.2 * |
1.5 * |
1.15e-08 |
4.33e-68 |
6.08e-02 |
0.0063 |
a33 |
EWSR1-FLI1_mut |
310 |
1 |
1 |
YM201636 |
FYV1 |
17 |
902 |
1.63 |
2.31 |
0.552 |
-1.76 |
1.4 * |
1.4 * |
1.5 * |
1.71e-08 |
2.74e-37 |
1.55e-02 |
0.0086 |
a35 |
EWSR1-FLI1_mut |
136 |
1 |
1 |
Mitomycin C |
DNA crosslinker |
17 |
856 |
2.77 |
-0.711 |
-2.31 |
-1.59 |
1 * |
1 * |
1.1 * |
3.19e-08 |
6.66e-06 |
1.12e-01 |
0.015 |
a42 |
EWSR1-FLI1_mut |
257 |
1 |
1 |
NPK76-II-72-1 |
PLK3 |
17 |
902 |
2.33 |
2.89 |
0.917 |
-1.98 |
1.2 * |
1.2 * |
1.6 * |
8.15e-08 |
1.57e-76 |
3.64e-02 |
0.032 |
a49 |
EWSR1-FLI1_mut |
228 |
1 |
1 |
KIN001-102 |
AKT1 |
17 |
905 |
2.33 |
2.63 |
0.892 |
-1.73 |
1.2 * |
1.2 * |
1.7 * |
2.17e-07 |
5.5e-67 |
1.74e-01 |
0.073 |
a55 |
EWSR1-FLI1_mut |
226 |
1 |
1 |
GSK1070916 |
AURKB |
15 |
879 |
0.94, 2.33 |
1.68 |
-1.29 |
-2.96 |
1.4 * |
1.4 * |
1.1 * |
1.09e-06 |
1.84e-60 |
3.56e-01 |
0.32 |
a56 |
EWSR1-FLI1_mut |
208 |
1 |
1 |
SB-715992 |
KIF11 |
17 |
905 |
-2.06 |
-1.74 |
-3.42 |
-1.68 |
0.95 . |
0.95 . |
1 * |
1.09e-06 |
6.99e-34 |
1.84e-02 |
0.32 |
a58 |
EWSR1-FLI1_mut |
221 |
1 |
1 |
TAK-715 |
p38a |
17 |
907 |
3.00, 2.33 |
4.19 |
3.29 |
-0.897 |
0.76 . |
0.76 . |
0.85 . |
1.22e-06 |
1.54e-54 |
2.29e-01 |
0.35 |
a59 |
EWSR1-FLI1_mut |
263 |
1 |
1 |
FR-180204 |
ERK |
17 |
902 |
3 |
4.92 |
3.99 |
-0.925 |
1.1 * |
1.1 * |
1.9 * |
1.37e-06 |
5.39e-37 |
7.83e-01 |
0.38 |
a61 |
EWSR1-FLI1_mut |
276 |
1 |
1 |
CAY10603 |
HDAC6 |
17 |
896 |
3 |
0.933 |
-0.52 |
-1.45 |
0.83 . |
0.83 . |
1.4 * |
1.43e-06 |
1.74e-36 |
6.16e-01 |
0.39 |
a68 |
EWSR1-FLI1_mut |
330 |
1 |
1 |
XMD13-2 |
RIPK |
17 |
903 |
2.33 |
3.39 |
2.03 |
-1.36 |
1 * |
1 * |
1.2 * |
2.78e-06 |
1.46e-57 |
7.47e-01 |
0.67 |
a70 |
EWSR1-FLI1_mut |
294 |
1 |
1 |
MPS-1-IN-1 |
MPS1 |
17 |
900 |
3 |
3.8 |
1.97 |
-1.84 |
1.4 * |
1.3 * |
1.9 * |
2.87e-06 |
9.63e-43 |
7.06e-01 |
0.68 |
a73 |
EWSR1-FLI1_mut |
1017 |
1 |
1 |
Olaparib |
PARP1, PARP2 |
16 |
831 |
1.61 |
3.56 |
2.03 |
-1.53 |
1.5 * |
1.5 * |
1.3 * |
3.05e-06 |
5.44e-21 |
2.22e-01 |
0.68 |
a76 |
EWSR1-FLI1_mut |
273 |
1 |
1 |
CUDC-101 |
HDAC, EGFR |
17 |
885 |
1.63 |
-0.264 |
-1.41 |
-1.15 |
0.65 . |
0.65 . |
1.3 * |
3.62e-06 |
1.1e-26 |
6e-01 |
0.79 |
a77 |
EWSR1-FLI1_mut |
345 |
1 |
1 |
KIN001-270 |
CDK9 |
17 |
902 |
3 |
4.89 |
4.15 |
-0.738 |
0.81 . |
0.81 . |
0.92 . |
3.81e-06 |
1.54e-57 |
5.26e-02 |
0.82 |
a78 |
EWSR1-FLI1_mut |
298 |
1 |
1 |
OSI-930 |
KIT, VEGFR, PDGFR |
17 |
903 |
2.33 |
4.03 |
3.11 |
-0.922 |
0.87 . |
0.87 . |
1.5 * |
3.93e-06 |
6.38e-59 |
2.76e-01 |
0.83 |
a102 |
EWSR1-FLI1_mut |
326 |
1 |
1 |
GSK690693 |
AKT |
17 |
902 |
2.33 |
3.6 |
1.66 |
-1.94 |
1.1 * |
1.1 * |
0.95 . |
1.33e-05 |
4.67e-50 |
8.03e-02 |
2.1 |
a107 |
EWSR1-FLI1_mut |
219 |
1 |
1 |
AT-7519 |
CDK9 |
17 |
902 |
2.33 |
1.2 |
-0.324 |
-1.53 |
0.77 . |
0.77 . |
1.2 * |
1.69e-05 |
4.09e-51 |
3.07e-01 |
2.6 |
a113 |
EWSR1-FLI1_mut |
256 |
1 |
1 |
JW-7-24-1 |
LCK |
17 |
904 |
2.33 |
1.62 |
0.126 |
-1.49 |
0.99 . |
0.99 . |
1.2 * |
1.81e-05 |
1.04e-47 |
9.06e-01 |
2.7 |
a114 |
EWSR1-FLI1_mut |
253 |
1 |
1 |
XMD14-99 |
EPHB3, CAMK1 |
17 |
904 |
2.33 |
4.38 |
3.26 |
-1.12 |
1.1 * |
1.1 * |
1.2 * |
1.87e-05 |
2.3e-73 |
6.42e-02 |
2.7 |
a115 |
EWSR1-FLI1_mut |
328 |
1 |
1 |
SNX-2112 |
HSP90 |
17 |
894 |
0.94 |
-0.541 |
-2.39 |
-1.85 |
0.86 . |
0.85 . |
1.4 * |
1.91e-05 |
2.54e-43 |
1.74e-01 |
2.7 |
a120 |
EWSR1-FLI1_mut |
272 |
1 |
1 |
AR-42 |
HDAC |
17 |
893 |
1.63 |
-0.251 |
-1.3 |
-1.05 |
0.61 . |
0.6 . |
1 . |
2.03e-05 |
1.99e-33 |
6.34e-01 |
2.8 |
a122 |
EWSR1-FLI1_mut |
303 |
1 |
1 |
PIK-93 |
PI4K, PI3K |
17 |
902 |
3 |
2.7 |
0.895 |
-1.81 |
1.1 * |
1.1 * |
1.2 * |
2.08e-05 |
9.86e-66 |
2.13e-01 |
2.8 |
a129 |
EWSR1-FLI1_mut |
252 |
1 |
1 |
WZ3105 |
CLK2, CNSK1E, FLT3, ULK1 |
17 |
904 |
0.247, 2.326 |
0.155 |
-1.54 |
-1.7 |
1 * |
1 * |
1.3 * |
2.39e-05 |
4.8e-58 |
9.63e-01 |
3.1 |
a131 |
EWSR1-FLI1_mut |
135 |
1 |
1 |
Gemcitabine |
DNA replication |
17 |
850 |
0.0237 |
-2.27 |
-4.65 |
-2.37 |
0.88 . |
0.88 . |
1.1 * |
2.54e-05 |
2.71e-08 |
1.33e-01 |
3.2 |
a144 |
EWSR1-FLI1_mut |
258 |
1 |
1 |
STF-62247 |
stimulates autophagy |
17 |
900 |
2.33 |
4.37 |
3.75 |
-0.619 |
0.63 . |
0.63 . |
0.81 . |
3.52e-05 |
9.71e-57 |
1.82e-01 |
4 |
a148 |
EWSR1-FLI1_mut |
274 |
1 |
1 |
PXD101, Belinostat |
HDAC |
17 |
876 |
3 |
0.305 |
-0.976 |
-1.28 |
0.68 . |
0.67 . |
1.2 * |
3.64e-05 |
4.62e-23 |
6.71e-01 |
4.1 |
a151 |
EWSR1-FLI1_mut |
299 |
1 |
1 |
OSI-027 |
MTORC1/2 |
16 |
899 |
3 |
1.15 |
-0.949 |
-2.1 |
0.92 . |
0.92 . |
1.1 * |
3.94e-05 |
1.93e-36 |
1e+00 |
4.3 |
a162 |
EWSR1-FLI1_mut |
346 |
1 |
1 |
THZ-2-102-1 |
CDK7 |
17 |
886 |
0.916, 2.996 |
-1.5 |
-3.97 |
-2.47 |
0.98 . |
0.98 . |
1.1 * |
5.13e-05 |
7.22e-38 |
3.51e-01 |
5.2 |
a165 |
EWSR1-FLI1_mut |
291 |
1 |
1 |
KIN001-266 |
MAP3K8 (COT) |
17 |
902 |
3 |
3.43 |
2.41 |
-1.02 |
0.9 . |
0.89 . |
1.4 * |
5.26e-05 |
7.76e-18 |
4.85e-01 |
5.2 |
a169 |
EWSR1-FLI1_mut |
288 |
1 |
1 |
KIN001-055 |
JAK3, MNK1 |
17 |
902 |
2.33 |
4.01 |
3.07 |
-0.944 |
0.91 . |
0.91 . |
1.2 * |
5.39e-05 |
2.11e-17 |
1.12e-01 |
5.3 |
a177 |
EWSR1-FLI1_mut |
304 |
1 |
1 |
SB52334 |
ALK5 |
17 |
901 |
3 |
4.25 |
2.5 |
-1.76 |
1.4 * |
1.4 * |
1.3 * |
6.04e-05 |
5.22e-23 |
5.26e-01 |
5.6 |
a179 |
EWSR1-FLI1_mut |
192 |
1 |
1 |
LFM-A13 |
BTK |
17 |
854 |
2.77 |
4.63 |
3.92 |
-0.713 |
0.98 . |
0.98 . |
1.1 * |
6.22e-05 |
6.09e-12 |
1.99e-01 |
5.7 |
a182 |
EWSR1-FLI1_mut |
1175 |
1 |
1 |
AG-014699 |
PARP1, PARP2 |
17 |
901 |
1.61 |
3.45 |
1.99 |
-1.46 |
1.5 * |
1.5 * |
1 * |
6.4e-05 |
7.74e-15 |
1.23e-01 |
5.8 |
a184 |
EWSR1-FLI1_mut |
225 |
1 |
1 |
Genentech Cpd 10 |
AURKA, AURKB |
17 |
906 |
2.33 |
2.54 |
0.603 |
-1.93 |
1.1 * |
1.1 * |
1.2 * |
6.47e-05 |
4.73e-42 |
3.08e-01 |
5.8 |
a186 |
EWSR1-FLI1_mut |
281 |
1 |
1 |
CH5424802 |
ALK |
17 |
901 |
1.63 |
3.67 |
2.58 |
-1.09 |
0.98 . |
0.98 . |
0.93 . |
6.76e-05 |
2.2e-29 |
8.41e-01 |
6 |
a198 |
EWSR1-FLI1_mut |
290 |
1 |
1 |
KIN001-260 |
IKK |
17 |
901 |
3 |
4.61 |
3.52 |
-1.09 |
0.91 . |
0.91 . |
1.2 * |
8.13e-05 |
2.77e-85 |
8.01e-01 |
6.8 |
a199 |
EWSR1-FLI1_mut |
305 |
1 |
1 |
TPCA-1 |
IKK |
17 |
902 |
3 |
3.02 |
1.48 |
-1.54 |
0.93 . |
0.92 . |
1.8 * |
8.28e-05 |
3.46e-62 |
7.83e-01 |
6.9 |
a203 |
EWSR1-FLI1_mut |
300 |
1 |
1 |
CX-5461 |
RNA Pol I |
17 |
900 |
2.33 |
3.03 |
0.894 |
-2.14 |
1.1 * |
1.1 * |
1 * |
8.99e-05 |
6.33e-45 |
2.24e-01 |
7.3 |
a223 |
EWSR1-FLI1_mut |
265 |
1 |
1 |
Tubastatin A |
HDAC6 |
17 |
899 |
3 |
4.41 |
3.36 |
-1.05 |
0.79 . |
0.79 . |
0.91 . |
1.16e-04 |
3.09e-74 |
7.65e-01 |
8.6 |
a245 |
EWSR1-FLI1_mut |
283 |
1 |
1 |
GSK2126458 |
PI3K, MTOR |
17 |
904 |
-2.0557, 0.0237 |
-2.92 |
-4.61 |
-1.69 |
0.95 . |
0.95 . |
0.9 . |
1.46e-04 |
9.76e-29 |
7.89e-01 |
9.9 |
a246 |
EWSR1-FLI1_mut |
152 |
1 |
1 |
CP466722 |
ATM |
17 |
906 |
2.77 |
2.64 |
1.39 |
-1.25 |
0.92 . |
0.91 . |
1.5 * |
1.51e-04 |
5.15e-45 |
3.29e-01 |
10 |
a250 |
EWSR1-FLI1_mut |
1248 |
1 |
1 |
FK866 |
NAMPT |
17 |
881 |
0 |
-2.51 |
-5.69 |
-3.19 |
1 * |
1 * |
1.5 * |
1.55e-04 |
6.28e-40 |
9.71e-04 |
10 |
a258 |
EWSR1-FLI1_mut |
223 |
1 |
1 |
ZSTK474 |
PI3K |
17 |
905 |
1.63, 2.33 |
1.03 |
-0.48 |
-1.51 |
0.89 . |
0.89 . |
0.99 . |
1.65e-04 |
5.76e-40 |
9.42e-01 |
11 |
a265 |
EWSR1-FLI1_mut |
1142 |
1 |
1 |
HG-5-113-01 |
LOK, LTK, TRCB, ABL(T315I) |
15 |
479 |
2.3 |
1.62 |
0.543 |
-1.08 |
1.1 * |
1.1 * |
1.8 * |
1.75e-04 |
9.25e-08 |
4.51e-01 |
11 |
a291 |
EWSR1-FLI1_mut |
222 |
1 |
1 |
BX-912 |
PDPK1 (PDK1) |
16 |
905 |
2.33 |
2.78 |
0.793 |
-1.99 |
1.1 * |
1.1 * |
1.3 * |
2.17e-04 |
4e-62 |
3.73e-01 |
12 |
a300 |
EWSR1-FLI1_mut |
287 |
1 |
1 |
KIN001-244 |
PDPK1 (PDK1) |
17 |
901 |
3 |
3.3 |
2.38 |
-0.92 |
0.73 . |
0.73 . |
0.9 . |
2.4e-04 |
2.83e-42 |
6.13e-01 |
13 |
a313 |
EWSR1-FLI1_mut |
341 |
1 |
1 |
EX-527 |
SIRT1 |
17 |
899 |
3 |
5.4 |
5.07 |
-0.324 |
0.49 |
0.49 |
0.76 . |
2.72e-04 |
5.9e-34 |
2.82e-01 |
14 |
a339 |
EWSR1-FLI1_mut |
268 |
1 |
1 |
YM155 |
BIRC5 (Survivin) |
17 |
871 |
1.63 |
-3.67 |
-5.16 |
-1.49 |
0.63 . |
0.62 . |
2 * |
3.16e-04 |
2.07e-09 |
3.96e-01 |
15 |
a345 |
EWSR1-FLI1_mut |
185 |
1 |
1 |
OSI-906 |
IGF1R |
17 |
851 |
0.94 |
2.12 |
-0.118 |
-2.24 |
1.6 * |
1.6 * |
1.3 * |
3.38e-04 |
1.67e-14 |
2.56e-01 |
16 |
a417 |
EWSR1-FLI1_mut |
331 |
1 |
1 |
QL-X-138 |
MNK2, PRKDC (DNAPK), MTOR, BTK, JAK3 |
17 |
891 |
2.33 |
1.58 |
0.146 |
-1.44 |
0.86 . |
0.86 . |
0.84 . |
5.09e-04 |
5.45e-40 |
6.73e-01 |
20 |
a432 |
EWSR1-FLI1_mut |
1494 |
1 |
1 |
SN-38 |
TOP1 |
18 |
909 |
-3 |
-4.19 |
-6.02 |
-1.83 |
1.1 * |
1.1 * |
0.96 . |
5.29e-04 |
2.82e-20 |
4.71e-03 |
20 |
a454 |
EWSR1-FLI1_mut |
301 |
1 |
1 |
PHA-793887 |
CDK-pan |
17 |
904 |
2.33 |
2.42 |
1.02 |
-1.4 |
0.68 . |
0.68 . |
0.67 . |
5.87e-04 |
6.81e-70 |
2.38e-01 |
21 |
a458 |
EWSR1-FLI1_mut |
271 |
1 |
1 |
VNLG/124 |
HDAC, RAR |
17 |
899 |
1.63 |
3.8 |
2.96 |
-0.839 |
0.87 . |
0.87 . |
0.85 . |
5.96e-04 |
2.07e-66 |
2.24e-01 |
21 |
a470 |
EWSR1-FLI1_mut |
134 |
1 |
1 |
Etoposide |
TOP2 |
17 |
865 |
2.77 |
1.6 |
-0.112 |
-1.71 |
0.88 . |
0.87 . |
0.92 . |
6.2e-04 |
2.54e-09 |
3.34e-02 |
22 |
a471 |
EWSR1-FLI1_mut |
1003 |
1 |
1 |
Camptothecin |
TOP1 |
16 |
830 |
-2.3 |
-4.01 |
-5.93 |
-1.92 |
1.1 * |
1.1 * |
1.2 * |
6.22e-04 |
3.45e-23 |
1.06e-01 |
22 |